Why model?
What If: Chapter 11
Elena Dudukina
2021-04-15
1 / 17
Welcome to Part II of Causal Inference Book


2 / 17
11.1 Data cannot speak for themselves
- A: anti-retroviral therapy
- Y: CD4 cell count at the end of the study
N: 16 individuals
Estimator: ^E[Y|A=a] (a function of the data) used to estimate the unknown populational parameter
- Consistent estimator: ^E[Y|A=a] satisfies the criterion with the increased sample size the estimate is closer to the populational value E[Y|A=a]
- Possible estimators:
- sample average of Y among those receiving A=a (a consistent estimator)
- the Y value of the first observation in the dataset with A=a (not a consistent estimator)
3 / 17
11.1 Data cannot speak for themselves
Population mean in the treated is the sample average 146.25 for those with A=1
Population mean in the untreated is the sample average 67.50 for those with A=0
Under exchangeability between A=1 and A=0, the average treatment effect (ATE) is 146.25−67.50=78.75
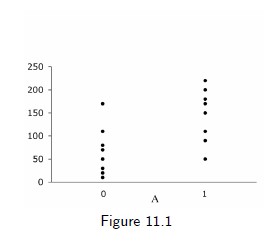
4 / 17
11.1 Data cannot speak for themselves
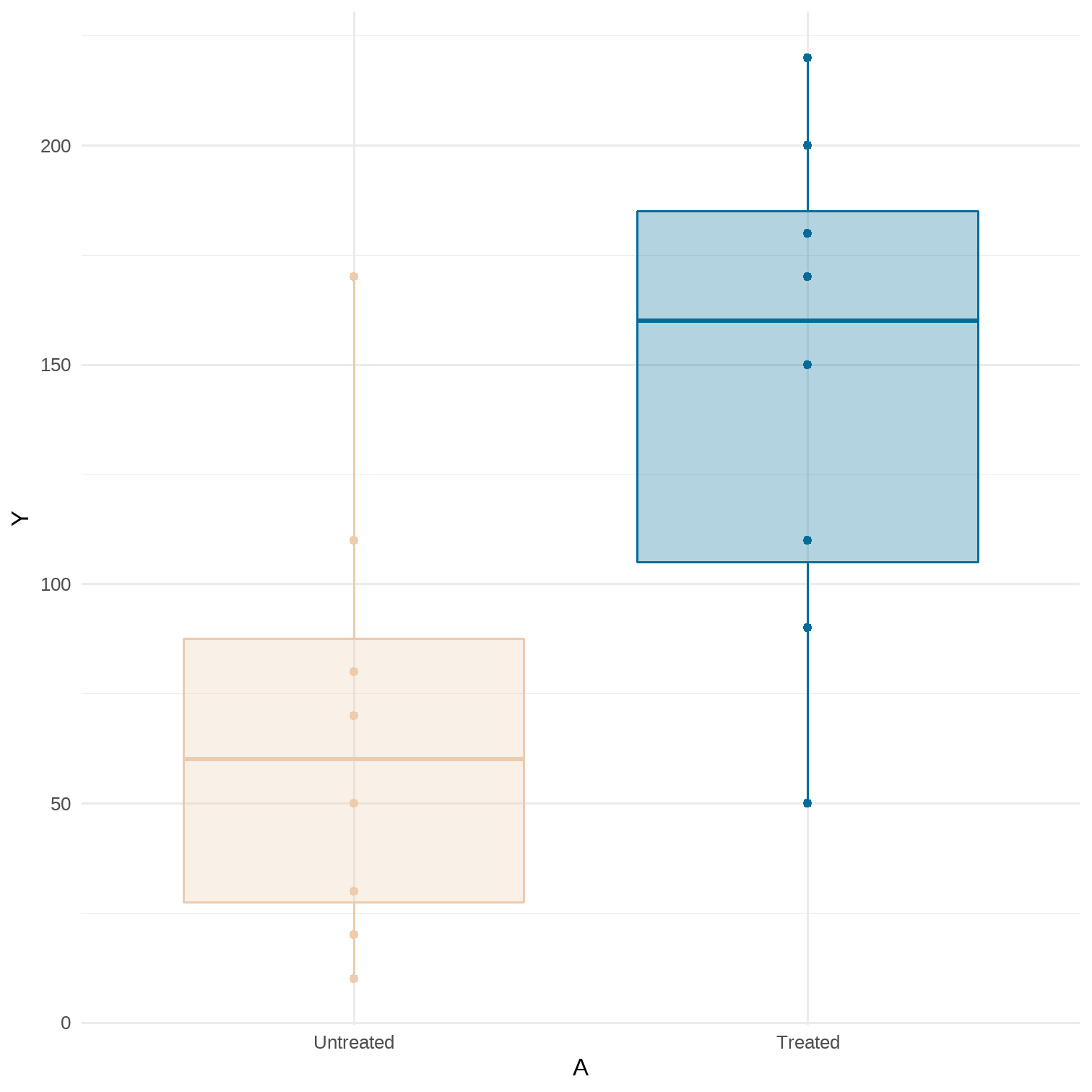
library(tidyverse)library(magrittr)# Sample averages by treatment level# Data for Figure 11.1A <- c(1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0)Y <- c(200, 150, 220, 110, 50, 180, 90, 170, 170, 30, 70, 110, 80, 50, 10, 20)data <- tibble(A, Y) %>% mutate(A = factor(A, levels = c("0", "1"), labels = c("Untreated", "Treated")))p <- data %>% ggplot(aes(x = A, y = Y, color = A, fill = A)) + geom_point() + geom_boxplot(alpha = 0.3) + theme_minimal() + theme(legend.position = "none") + scale_color_manual(values = wesanderson::wes_palette(name = "Darjeeling2", n = 2)) + scale_fill_manual(values = wesanderson::wes_palette(name = "Darjeeling2", n = 2))data %>% group_by(A) %>% summarise(mean = mean(Y)) %>% kableExtra::kable()| A | mean |
|---|---|
| Untreated | 67.50 |
| Treated | 146.25 |
5 / 17
11.1 Data cannot speak for themselves
A is polytomous variable
- no treatment (A = 1)
- low-dose treatment (A = 2)
- medium-dose treatment (A = 3)
- high-dose treatment (A = 4)
Probability of getting any treatment level is 0.25
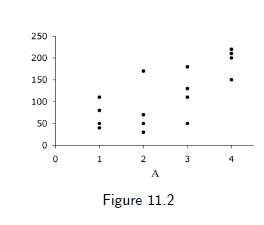
6 / 17
11.1 Data cannot speak for themselves
# Sample averages by treatment level# Data for Figure 11.2A <- c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4)Y <- c(110, 80, 50, 40, 170, 30, 70, 50, 110, 50, 180, 130, 200, 150, 220, 210)data <- tibble(A, Y) %>% mutate(A = factor(A))p <- data %>% ggplot(aes(x = A, y = Y, color = A, fill = A)) + geom_point() + geom_boxplot(alpha = 0.3) + theme_minimal() + theme(legend.position = "none") + scale_color_manual(values = wesanderson::wes_palette(name = "Darjeeling1", n = 4)) + scale_fill_manual(values = wesanderson::wes_palette(name = "Darjeeling1", n = 4))data %>% group_by(A) %>% summarise(mean = mean(Y)) %>% kableExtra::kable()| A | mean |
|---|---|
| 1 | 70.0 |
| 2 | 80.0 |
| 3 | 117.5 |
| 4 | 195.0 |
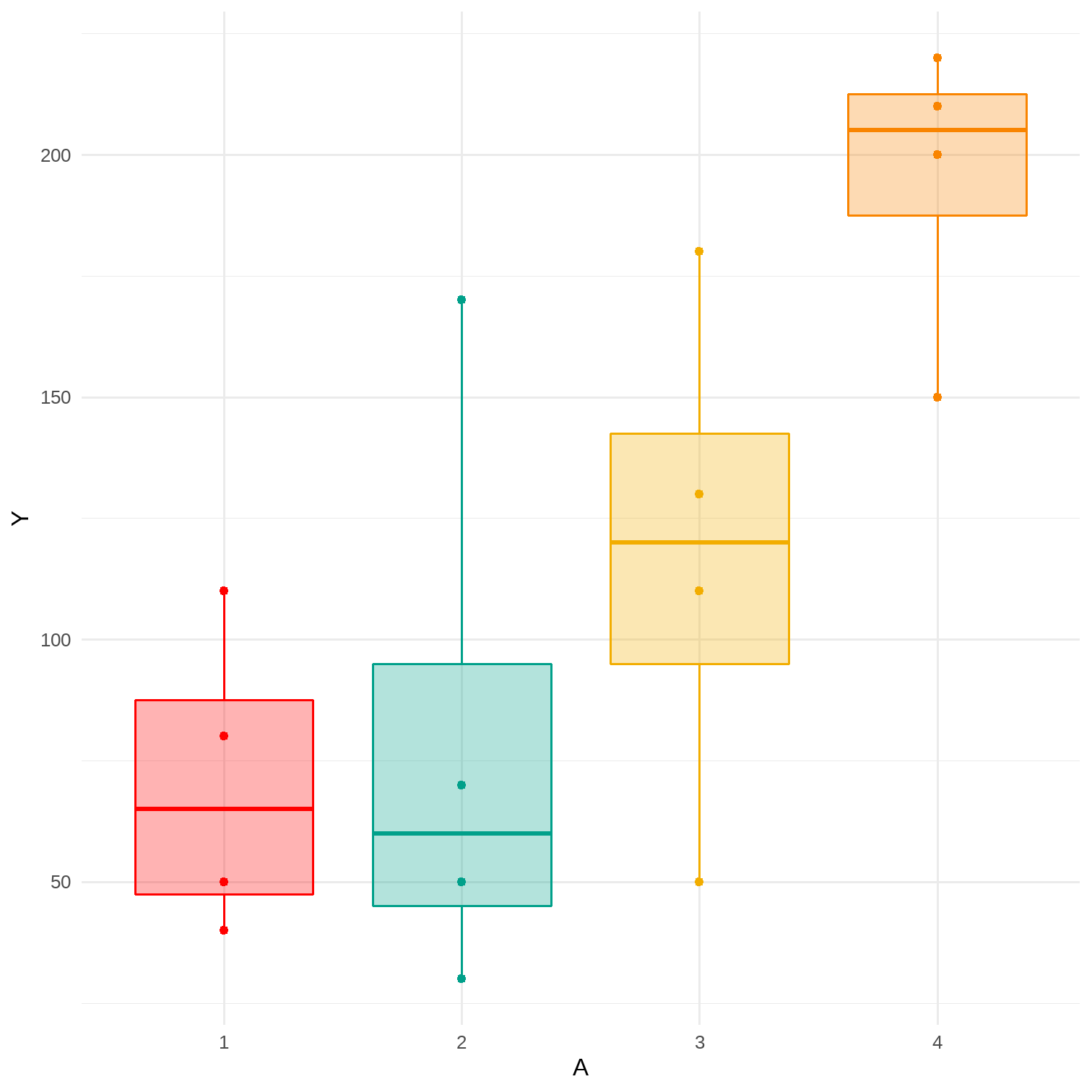
7 / 17
11.1 Data cannot speak for themselves
- A is a dose treatment in in mg/day
- Values [0;100]
- A continuous variable is a categorical variable with infinite number of categories
- estimate, in the target population, the mean of the outcome Y among individuals with treatment level A = 90
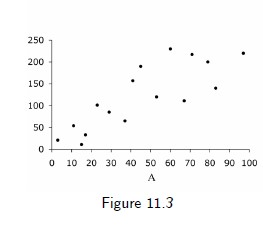
8 / 17
11.1 Data cannot speak for themselves
# 2-parameter linear model# Data for Figures 11.3A <- c(3, 11, 17, 23, 29, 37, 41, 53, 67, 79, 83, 97, 60, 71, 15, 45)Y <- c(21, 54, 33, 101, 85, 65, 157, 120, 111, 200, 140, 220, 230, 217, 11, 190)data <- tibble(A, Y)rm(A, Y)res_lm <- lm(Y ~ A, data = data) %>% broom::tidy(., conf.int = T) %>% select(1, 2, 6, 7)p <- data %>% ggplot(aes(x = A, y = Y)) + geom_point() + theme_minimal()## # A tibble: 2 x 4## term estimate conf.low conf.high## <chr> <dbl> <dbl> <dbl>## 1 (Intercept) 24.5 -21.2 70.3 ## 2 A 2.14 1.28 2.99
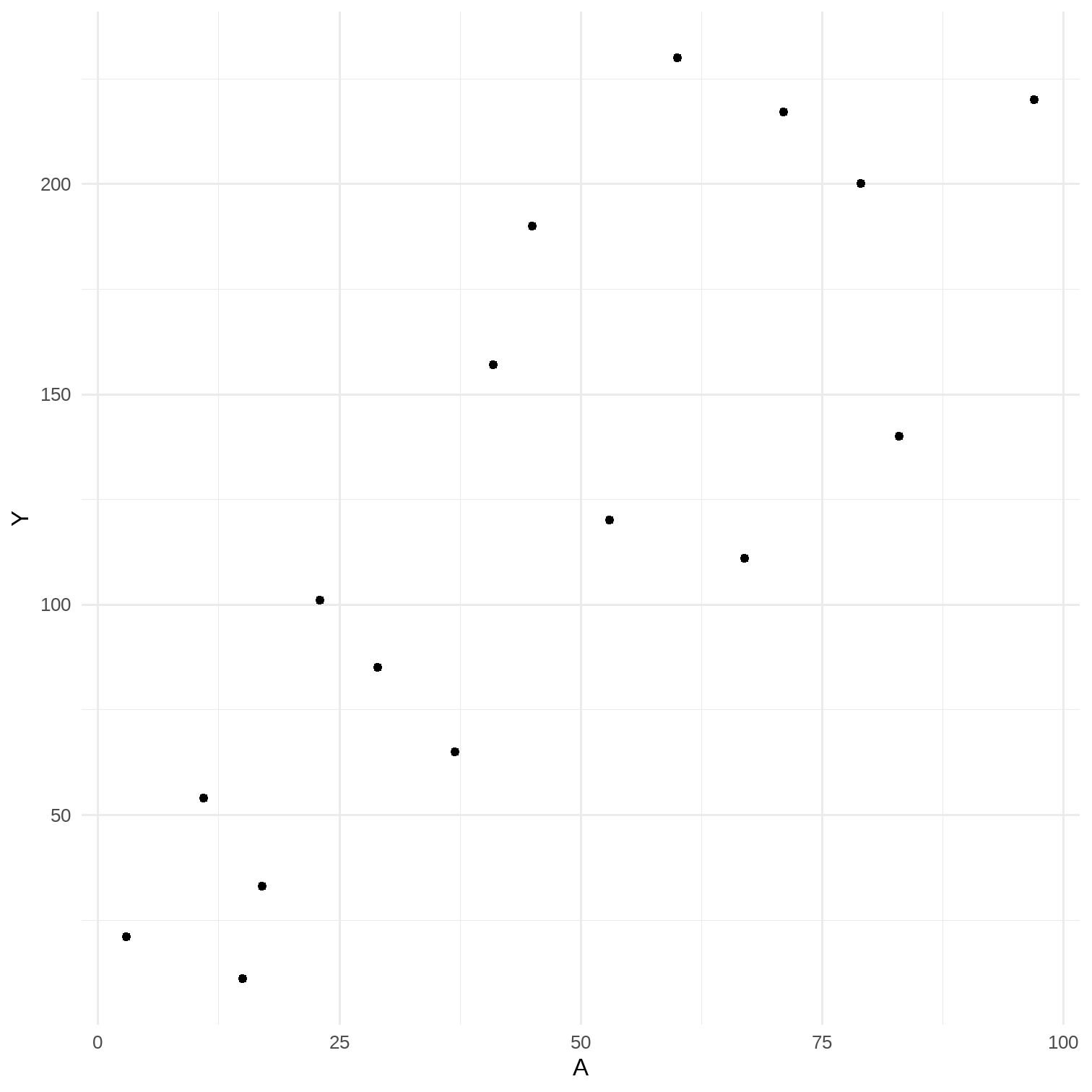
9 / 17
11.2 Parametric estimators of the conditional mean
- Aim: to estimate mean of Y among individuals with treatment level A = 90, or E[Y|A=90]
- Y ~ Normal(A, ϵ) (Y is a function of A with some error term)
- The mean of Y changes from some value θ0 by θ1 units per unit of treatment A: E[Y|A]=θ0+θ1A
- The shape of conditional mean E[Y|A] is determined by this equation - linear mean model
- θ0 and θ1 are parameters of the model
- If model describes the expectation with a finite number of parameters, the model is parametric
10 / 17
# Figure 11.4p <- data %>% ggplot(aes(x = A, y = Y)) + geom_point() + geom_smooth(method = lm, color = "#00868B") + theme_minimal()p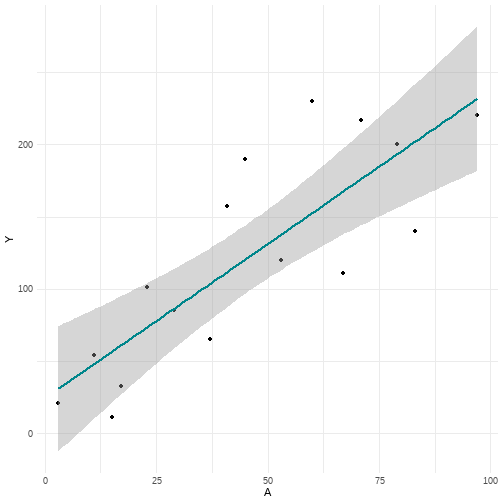
lm(Y ~ A, data = data) %>% broom::tidy(., conf.int = T) %>% select(1, 2, 6, 7)## # A tibble: 2 x 4## term estimate conf.low conf.high## <chr> <dbl> <dbl> <dbl>## 1 (Intercept) 24.5 -21.2 70.3 ## 2 A 2.14 1.28 2.9924.546369 + 2.137152*90## [1] 216.8911 / 17
11.2 Parametric estimators of the conditional mean
- A model restricts the joint distribution of the data
- Parametric models come with the assumptions
- The inferences are valid only when the model is correctly specified
- Assumption of no model misspecification for the model-based causal inference
12 / 17
11.3 Nonparametric estimators of the conditional mean
For dichotomous treatment A:
- E[Y|A]=θ0+θ1A
- E[Y|A=1]=E[Y|A=0]+θ1
- Saturated model
- "Model is saturated whenever the number of parameters in a conditional mean model is equal to the number of unknown conditional means in the population"
- "When a model has only a few parameters but it is used to estimate many population quantities, it is parsimonious"
13 / 17
11.4 Smoothing
- Linear model with quadratic term A2 (or other polynomials: ..., A15)
- E[Y|A]=θ0+θ1A+θ2A2
- The more parameters the model has, the less smooth the curve is
14 / 17
11.4 Smoothing
data %<>% mutate(A_sq = A*A)lm(Y ~ A + A_sq, data = data) %>% broom::tidy(., conf.int = T) %>% select(1, 2, 6, 7)## # A tibble: 3 x 4## term estimate conf.low conf.high## <chr> <dbl> <dbl> <dbl>## 1 (Intercept) -7.41 -76.0 61.2 ## 2 A 4.11 0.800 7.41 ## 3 A_sq -0.0204 -0.0535 0.0127# predict by hand-7.40687745 + 4.10722663*90 -0.02038477*90^2## [1] 197.1269# 3 parametersp <- data %>% ggplot(aes(x = A, y = Y)) + geom_point() + theme_minimal() + stat_smooth(method = "glm", formula = y ~ poly(x, 2), color = "#00868B")# 7 parametersp2 <- data %>% ggplot(aes(x = A, y = Y)) + geom_point() + theme_minimal() + stat_smooth(method = "glm", formula = y ~ poly(x, 6), color = "#00868B")
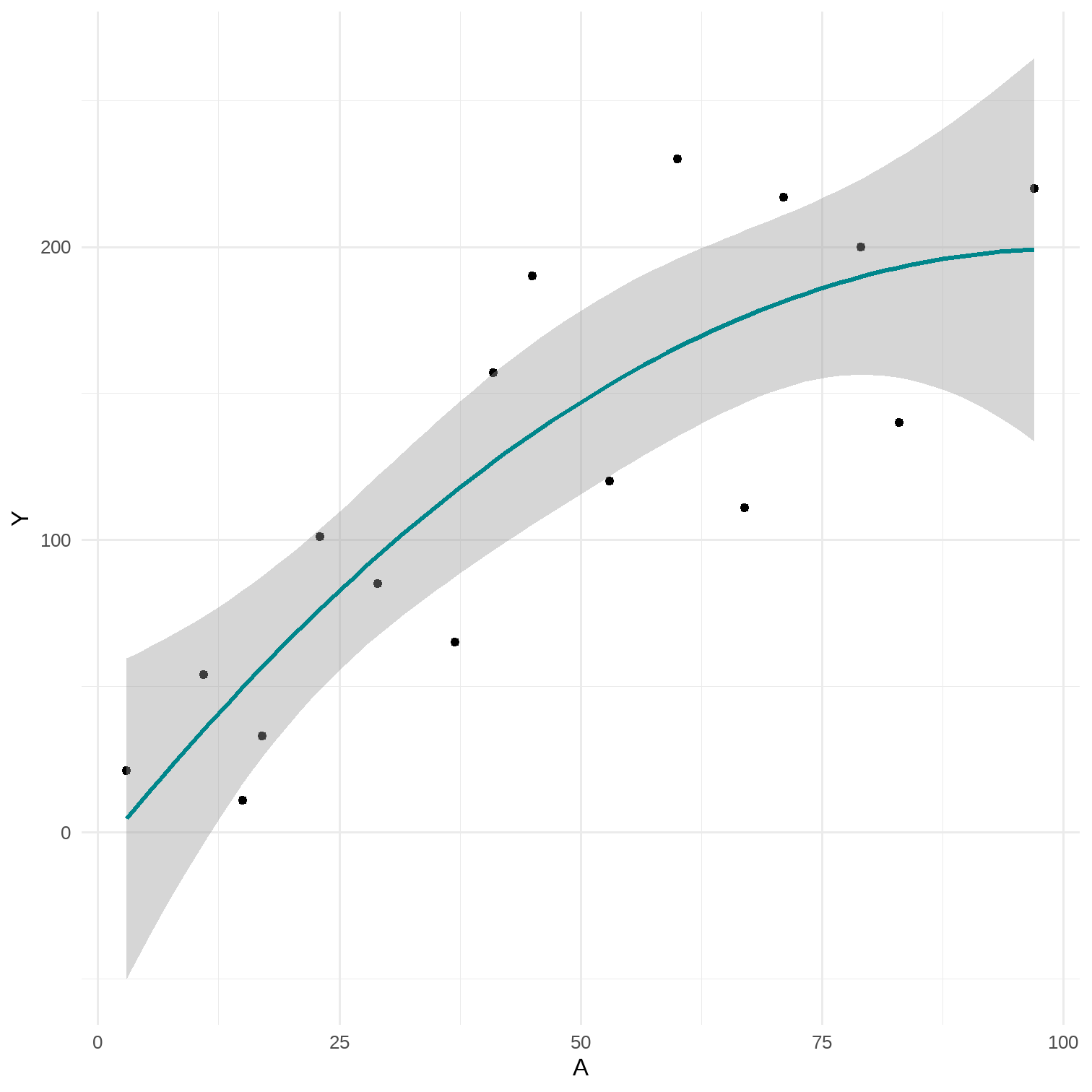
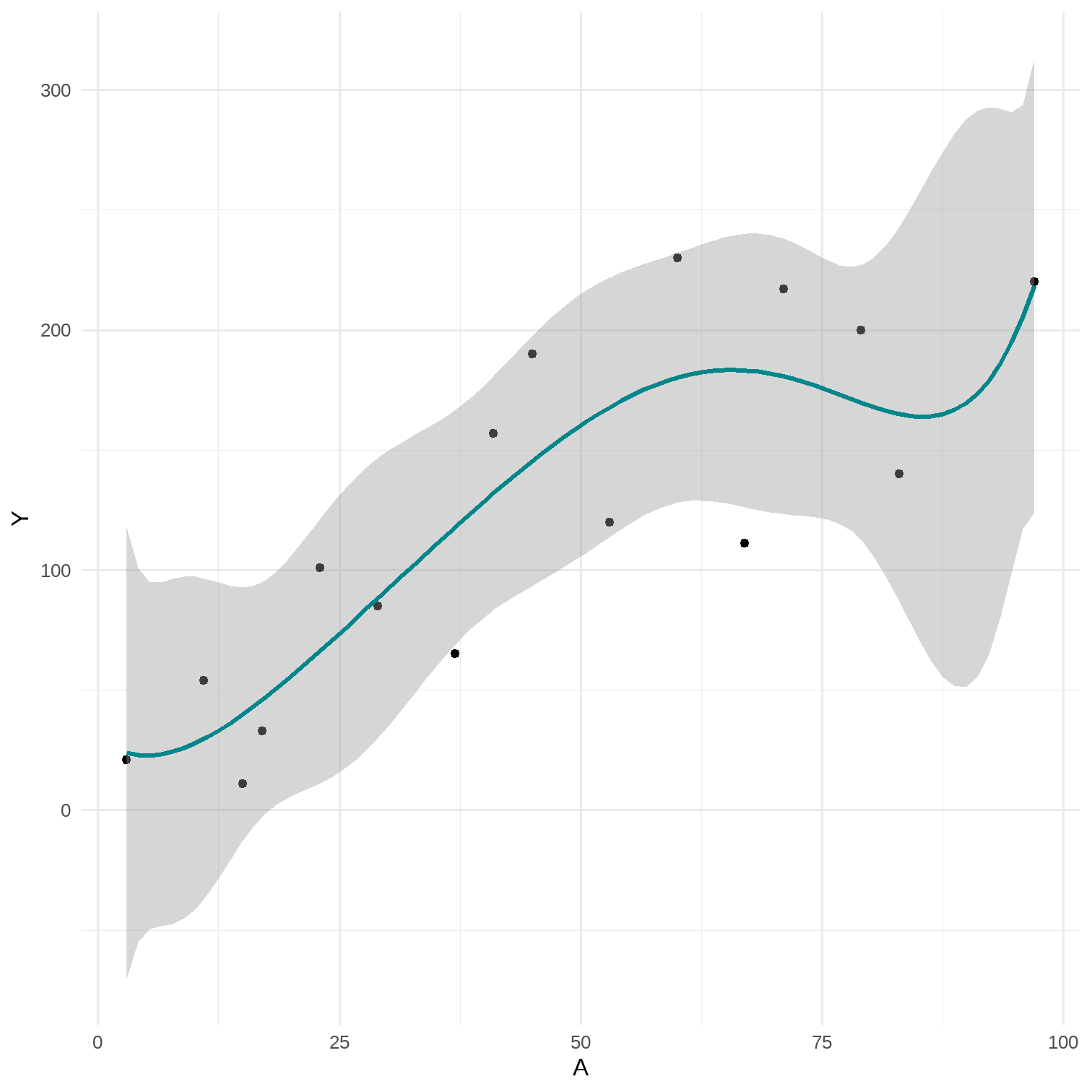
15 / 17
11.5 The bias-variance trade-off
- Under 2-parameter model the prediction for CD4 cell count given A=90 was 216.9 and under 3-parameter model it was 197.1
- 3-parameter model is correctly specified under both straight line and curvelinear scenarios
- More parameters, less restrictions model implies
- Less smooth models provide less biased, but more imprecise result (estimate with a larger variance)
16 / 17
References
Hernán MA, Robins JM (2020). Causal Inference: What If. Boca Raton: Chapman & Hall/CRC (v. 31mar21)
17 / 17